Ligand/Receptor Interactions in Malaria Research
- Malaria remains a global killer infecting upwards of 260 million people annually and leading to over 590,000 deaths in 2023 (World Health Organization Annual Malaria Report).
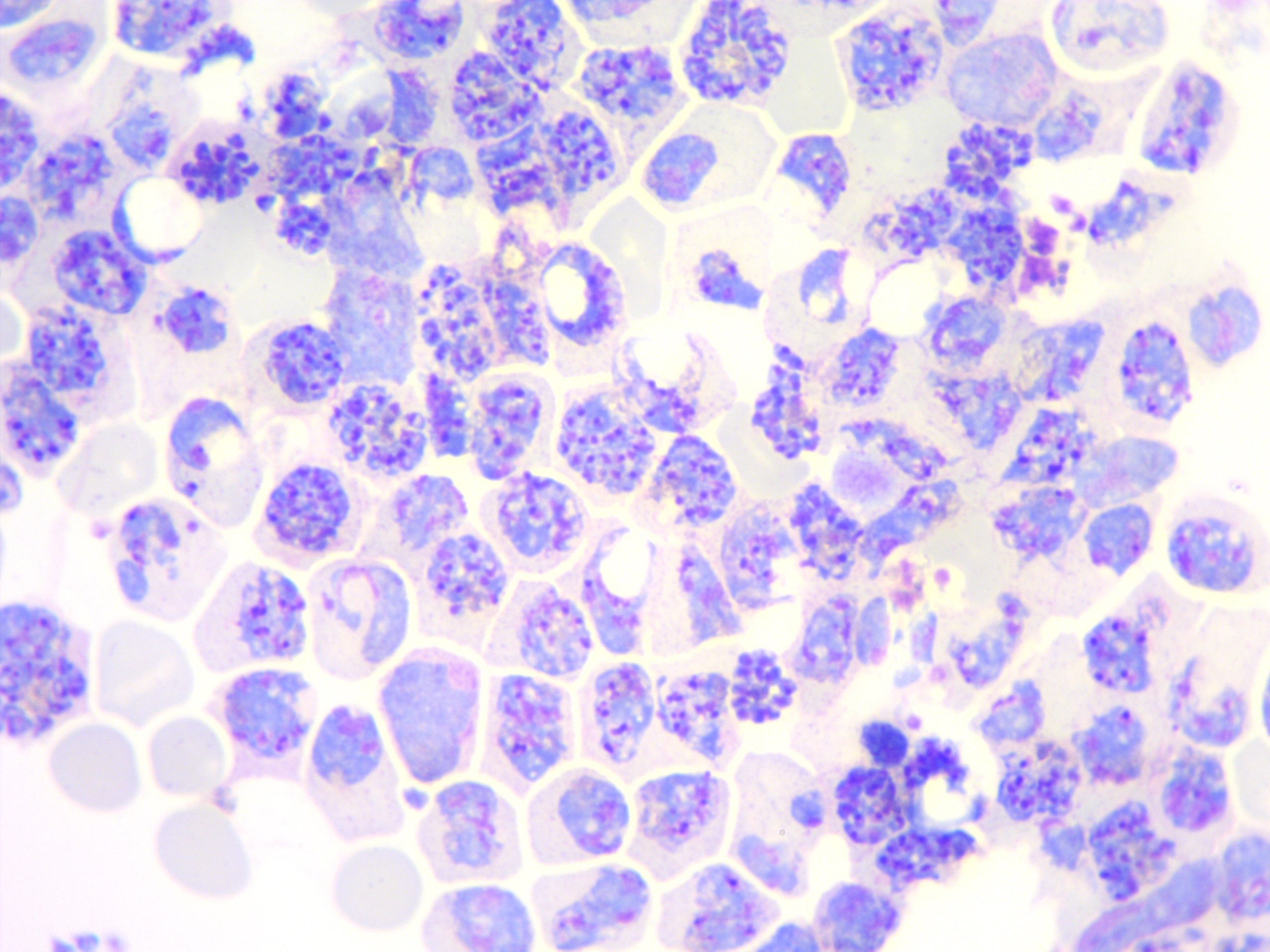
- Malaria is caused by the single-celled Apicomplexan parasite transmitted by the bite of an infected female Anopheles mosquito. Six species of Plasmodium parasites cause malaria in humans.
- Parasite ligand/host receptor interactions are essential for mediating multiple aspects of parasite life cycle including parasite invasion into red blood cells (RBCs), rosetting and cytoadherence.
- Parasites have evolved a large number of ligand proteins and identifying essential interactions is key to identifying therapeutic targets.

RBC genetic systems to identify host factors for parasite invasion
- In vitro culture and differentiation of either primary (CD34+ hematopoietic stem cell – cRBCs) or immortalized erythroid cell lines into orthochromatic cells or reticulocytes recapitulates key aspects of erythropoiesis
- Plasmodium spp. parasites are capable of infecting cRBCs or immortalized cell lines, suggesting that these cells are good model systems for understanding parasite invasion
- cRBCs and immortalized cell lines support genetic modification (CRISPR/Cas9, RNAi)

Genetic screening to identify and validate host factors required for parasite invasion
- Forward genetic screens of pooled RBC membrane proteins are used to identify candidate host receptors
- Reverse genetics and complementation approaches can be used to validate new candidate host receptors
- Candidate host receptors can be identified via population genetic studies